This project was a mix of challenges and learning as I navigated the CPython C API and worked closely with the NumPy community. I want to share a behind-the-scenes look at my work on introducing a new string DType in NumPy 2.0, mostly drawn from a recent talk I gave at SciPy. In this post, I’ll walk you through the technical process, key design decisions, and the ups and downs I faced. Plus, you’ll find tips on tackling mental blocks and insights into becoming a maintainer.
By the end, I hope you’re going to have the answers to these questions:
First, I’ll start with a brief history of strings in NumPy to explain how strings worked before NumPy 2.0 and why it was a little bit broken.
"strings" were what we now call byte strings in Python 3 – arrays of arbitrary bytes with no attached encoding. NumPy string arrays had similar behavior.
Python 2.7.18 (default, Jul 1 2024, 10:27:04)
>>> import numpy as np
>>> np.array(["hello", "world"])
array(['hello', 'world'], dtype='|S5')
>>> np.array(['hello', '☃'])
array(['hello', '\xe2\x98\x83'], dtype='|S5')
Let’s say you create an array with the contents “hello", "world”, you can see it gets created with the DType “S5”. So, what does that mean? It means it’s a Python 2-string array with five elements, five characters, or five bytes per array (characters and bytes are the same thing in Python 2).
'hello', ‘☃’ and if you happen to know the UTF-8 bytes for Unicode 'snowman', it’s '\xe2\x98\x83'. So, it’s just taking the UTF-8 bytes from my terminal and putting them straight into the array. ![Diagram illustrating the memory layout of a NumPy string array. It shows two elements in the array: the first element contains the characters 'h', 'e', 'l', 'l', and 'o', and the second element contains encoded bytes represented as '\xe2', '\x98', '\x83', followed by two null bytes 'b\x00'. The array elements are labeled as 'arr[0]' and 'arr[1]'.](https://quansight.com/wp-content/uploads/2024/10/image1.png)
Here, we have the bytes in the Python 2 string array: the ASCII byte for 'h', the ASCII byte for 'e', and over in the second element of the array is the UTF-8 bytes for the Unicode snowman. It’s also important to know that for these fixed-width strings—if you don’t fill up the width of the array, it just adds zeros to the end, which are null bytes.
>>> arr = np.array([u'hello', u'world'])
>>> arr
array([u'hello', u'world'], dtype='<U5')
Python 2 also had this Unicode type, where you could create an array with the contents 'hello', 'world', but as Unicode strings, and that creates an array with the DType 'U5'. This works, and it’s exactly what Python 2 did with Unicode strings. Each character is a UTF-32 encoded character, so four bytes per character
![Diagram showing the memory layout of a NumPy string array using UTF-32 encoding. It displays two elements: 'arr[0]' contains the characters 'h', 'e', 'l', 'l', 'o', and 'arr[1]' contains the characters 'w', 'o', 'r', 'l', 'd'. Each character is represented with a prefix 'u' indicating Unicode representation. A detailed inset illustrates the UTF-32 encoding for the character 'h', represented as 'u'h'' with its corresponding byte sequence 'b'h\x00\x00\x00'.](https://quansight.com/wp-content/uploads/2024/10/image2.png)
>>> arr = np.array(['hello', 'world'])
>>> arr
array(['hello', 'world'], dtype='<U5')
>>> arr.tobytes()
b'h\x00\x00\x00e\x00\x00\x00l\x00\x00\x00l\x00\x00\x00o\x00\x00\x00w\x00\x00\x00o\x00\x00\x00r\x00\x00\x00l\x00\x00\x00d\x00\x00\x00'
To make matters worse, string operations were slow, too. In a blog post that a colleague of mine at Quansight Labs, Lysandros Nikolaou, wrote, he did a project where he rewrote the loops for UFunc operations using the fixed-width string DTypes (the “S” and “U” DTypes I mentioned earlier).

>>> arr = np.array(
['this is a very long string', np.nan, 'another string'],
dtype=object
)
>>> arr
array(['this is a very long string', nan, 'another string'],
dtype=object)
dtype=object and it stores the Python strings and Python objects that you put into the array directly. These are references to Python objects. If we call np.isnan on the second element of the array, you get back np.True_ because the object is np.nan, and the other elements are python strings stored directly in the array.
>>> arr = np.array(
['this is a very long string', np.nan, 'another string'],
dtype=object
)
>>> np.isnan(arr[1])
np.True_
>>> type(arr[0])
str
However, the choice to go with object arrays as the default for string arrays is basically ecosystem-wide technical debt. For example, this has had a significant impact on pandas. If we create a pandas DataFrame with one column of names and look at that column, we will see that it’s actually backed by a NumPy object array. This has caused an enormous amount of pain for pandas over the years because object arrays are slow – somewhat of an unloved feature that can’t possibly work as efficiently or seamlessly as the rest of NumPy.

There was a proposal to replace the object string arrays with PyArrow strings (PDEP-10), which was accepted. However, since PyArrow is a big dependency, they are now trying to decide whether or not to make PyArrow a required dependency, and PDEP-14 and PDEP-15 are follow-ups to this. It’s all mixed up, and there are now four or five different string DTypes in pandas, which is just kind of a mess.
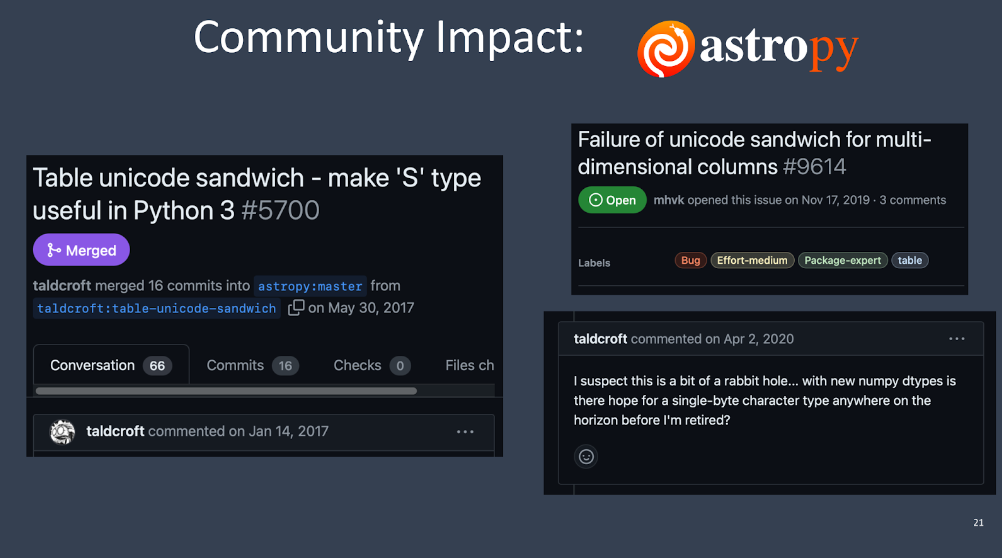
Another community impact example is Astropy. Astropy has something inside it called the Unicode Sandwich. It generated a pull request with 66 comments. There is another issue that opened two years after this feature was added, and the person who wrote the original pull request commented, “I don’t know how to fix this. Why can’t NumPy just fix itself?” Astropy had to do a lot to work around NumPy’s bad Unicode string default.
The NumPy project was very much aware of this. In 2017, Julian Taylor proposed to the mailing list that the Python 3 four-byte UTF-32 encoding wasn’t great and something should be done about it. This generated a lot of discussion (As you can see below).

Out of this discussion, we added the need for a new string DType, something that works sort of like 'dtype=object' but is type-checked to the NumPy roadmap. We also discussed adding a variable-width string DType with UTF-8 encoding.
Three years later, not much had happened inside NumPy to fix the string DType situation, but lots of work was happening to improve NumPy’s DType infrastructure and make it possible to solve this issue and many other issues that could be solved by defining a new DType. When that work was starting to shape up in late 2019, when NASA had a funding opportunity through the ROSES Grant, many projects across the ecosystem saw an opportunity to fix long-standing issues. Pandas, Scikit-learn, SciPy, and NumPy collectively proposed a grant with Quansight Labs leading it. Cal Poly San Luis Obispo, through Matt Haberland, and Los Alamos National Labs, through Tyler Reddy, were also involved. I’m not going to go through everything we did in this grant, but these were all big, fundamental improvements to these libraries. A lot of the maintenance for these libraries has been paid for out of these grants for the past few years. One of the things proposed in this grant was a variable-width string DType for NumPy.
I want to back up and talk about myself for a minute. I have a PhD in astrophysics from UC Santa Cruz, where I did simulations of galaxies on supercomputers. The technical skills I learned from that were 1990s-style C++ and Python. After that, I did a postdoc and later became a research scientist at the University of Illinois, where I worked as a maintainer for the yt project. There, I learned a lot of scientific Python skills (particularly working with Cython) and open source maintainership skills. All that to say, when I came to NumPy, I wasn’t coming in from zero with no systems programming or maintainer experience.
After leaving academia in 2019, I did a batch at the Recurse Center, where I worked on some Rust projects, so I have some Rust experience as well. Then, I came to work at Quansight, where I mostly worked on PyTorch. I learned a lot of modern C++ and pybind11 there, gaining some skills, though not necessarily a 100% overlap with what was needed for this project
How do you get started on a big project like that, and how do you get help? Often, projects will have public channels where you can ask questions. NumPy has a Slack channel. Anybody can join, and anyone can ask questions. Often, projects will have some kind of channel like that—use it, that’s what it’s for.
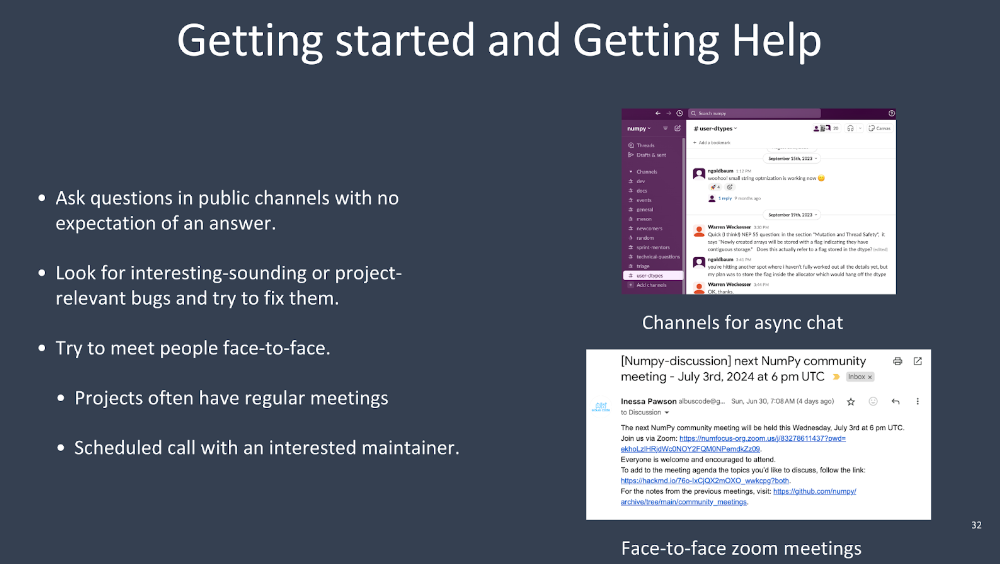
Another nice thing is to look for interesting or project-relevant bugs—bugs that might be related to the code you’re going to touch later for the project. Try to fix them to get acclimated to the codebase. Another great thing that NumPy has, which I think other projects should adopt if they don’t, is regular face-to-face Zoom meetings. It’s so important to change someone from just a GitHub handle communicating through text to a human face with emotions. So many conflicts can be diffused just by talking to someone.

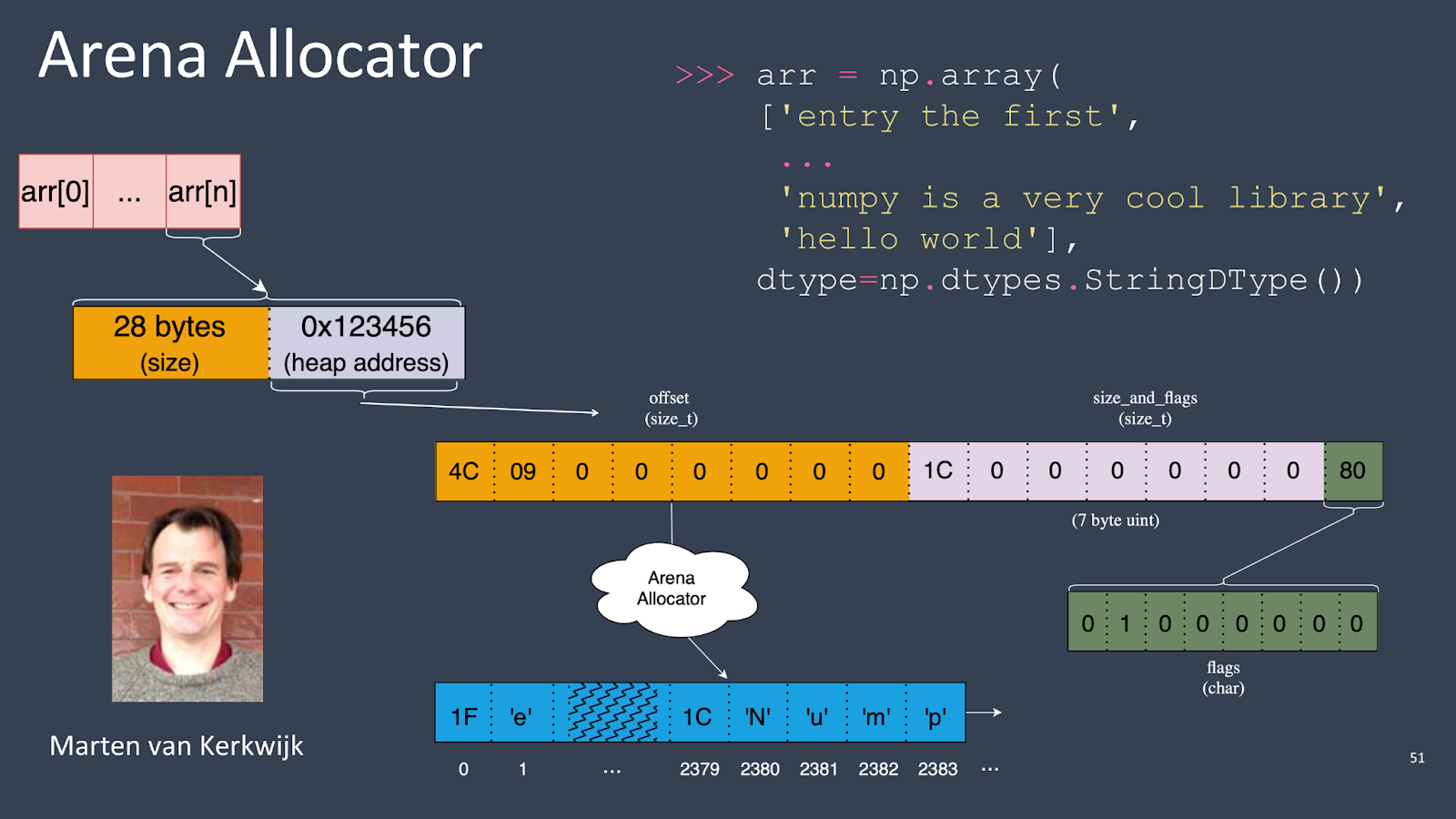
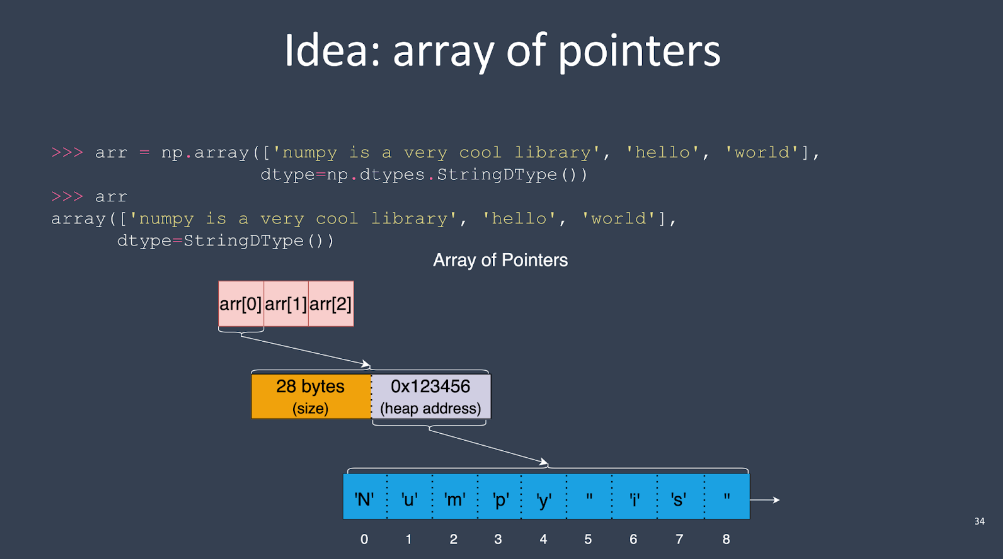
But with variable-width strings, you have to do something else unless you want to store the string data in two memory buffers, one storing the strings and another storing the offsets into the string data where each array element starts. The offsets array can be accessed just like a normal NumPy array.
The problem with that approach is that NumPy doesn’t offer a way to store data outside of the array buffer—there’s no concept of “sidecar storage” in NumPy.
To actually build it, we decided to make a prototype. It’s good to work in a public repository but not necessarily in the main project repository since exploratory work can be noisy and attract unnecessary attention from developers who aren’t working on your project. People who are interested in the project can still subscribe to your repo, and you can still do code review, but it may not be as rigorous as if it were a real contribution. Making the barrier to commit code low is critical for exploratory coding like this.

I also highly recommend Julia Evans’ zine on debugging. If you’re looking for a resource to improve your debugging skills, start there.

How did I become part of that community? Becoming a maintainer is about becoming useful to the project. One thing people often don’t realize is that anybody can review code. You don’t have to be a maintainer to review someone else’s pull request. In fact, a great way to become a maintainer is by starting to review code. A good rule of thumb is for every pull request you submit, review one other pull request. This helps unblock the project since one of the main things slowing progress is a lack of code reviews.
npy_string_dtype. 
In NumPy 2.0, there is a new string DType available: 'np.dtype.StringDType'. It supports UTF-8 encoded variable-width strings. For example, you can have a Unicode emoji, and it’s reproduced exactly. This DType can store strings in NumPy 2.0, so when we call 'str_len()' on the string, it gives the correct answer. Keep in mind that 'str_len()' returns the length in bytes. It is, in fact, a ufunc. The string DType in NumPy 2.0 handles these new string formats, and it works with all the standard NumPy DTypes.
>>> arr = np.array(
... ["this is a very long string: 😎", "short string"],
... dtype=StringDType())
>>> arr
array(['this is a very long string: 😎', 'short string'],
dtype=StringDType())
>>> np.strings.str_len(arr)
array([29, 12])
>>> isinstance(np.strings.str_len, np.ufunc)
True
One of the significant improvements is that it now supports missing data directly. You can create a string DType with this ‘na’ object parameter. If you specify the 'nan' object, then the DType can represent missing data directly in the array. For example, you can call 'np.isnan()' on an array, and it will return 'True' for any entries that are 'nan'. This is one of the major reasons why people preferred object string arrays over NumPy’s existing string data types. Adding first-class support for missing data in StringDType makes it much easier to transition existing codebases relying on object string arrays to use StringDType arrays.
>>> dt = StringDType(na_object=np.nan)
>>> arr = np.array(["hello", nan, "world"], dtype=dt)
>>> arr[1]
nan
>>> np.isnan(arr[1])
True
>>> np.isnan(arr)
array([False, True, False])
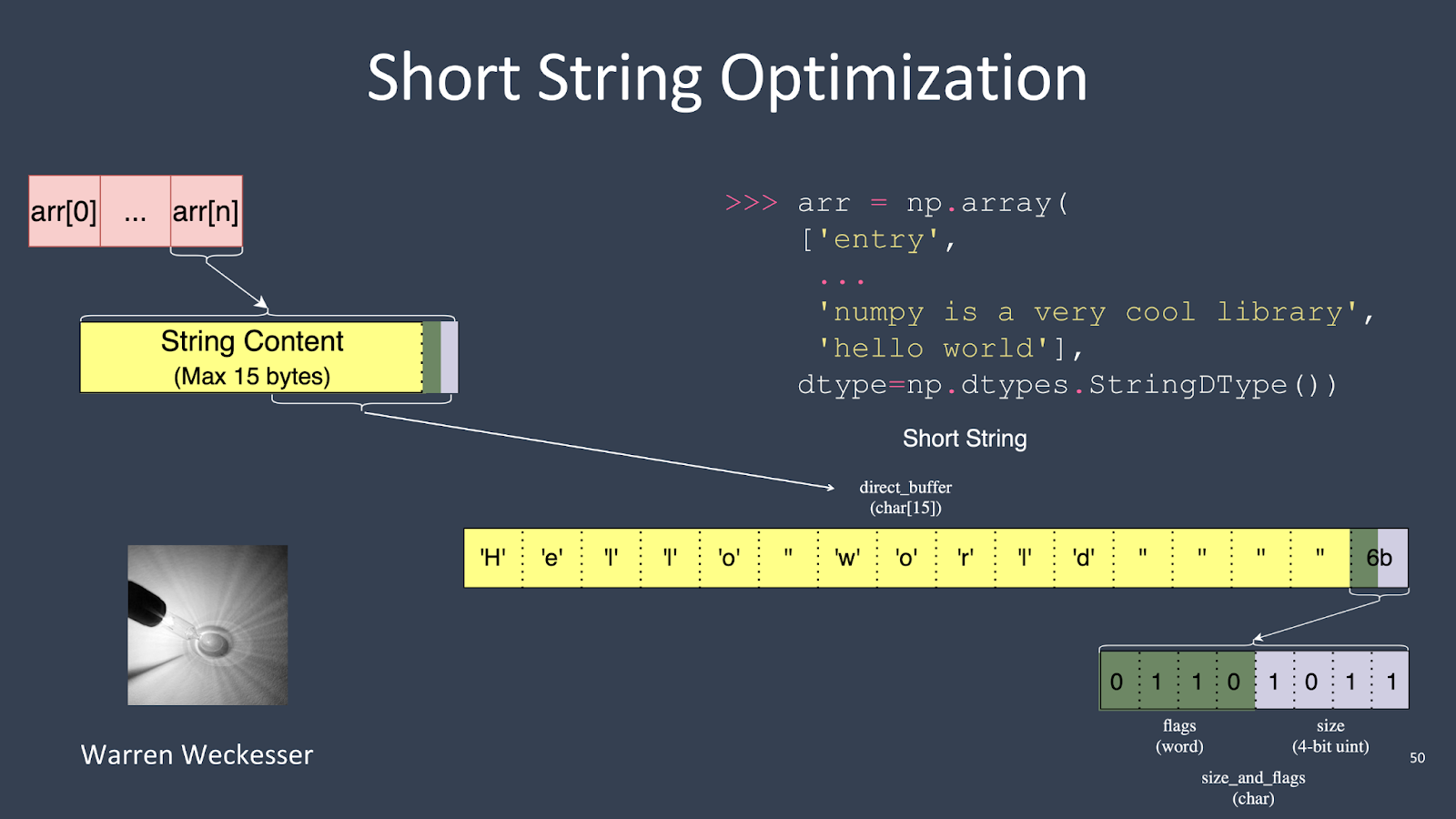
Another cool feature is the short string optimization. For instance, if you have an array and the last entry is ‘hello, world’, which only needs 11 bytes to store, the struct that holds the entry in the array is 16 bytes. So, instead of doing a separate heap allocation, NumPy can store small strings directly in the array buffer. This optimization avoids unnecessary heap allocations for smaller strings.
NumPy