cuCIM is a new RAPIDS library for accelerated n-dimensional image processing and image I/O. The project is now publicly available under a permissive license (Apache 2.0) and welcomes community contributions. This is the second part of a joint blog post with NVIDIA. Both posts feature a common motivation section, but the NVIDIA post focuses on cuCIM software architecture, image I/O functionality, and benchmark results. In this post, we expand on the CuPy-based cucim.skimage package, which provides a CUDA-based implementation of the scikit-image API. We will give an overview of how existing CPU-based scikit-image code can be ported to the GPU. We will also provide guidance on how to get started using and contributing to cuCIM. The initial release of the library was a collaboration between Quansight and NVIDIA’s RAPIDS and Clara teams.
As a quick first look, the following code applies a set of filters that highlight vessels in an image of the retina.
import cupy as cp
import numpy as np
import matplotlib.pyplot as plt
from skimage import data
from cucim.skimage import color
from cucim.skimage import filters
# load example data and transfer it to the GPU
retina = data.retina()
retina_gpu = cp.asarray(retina)
# convert color image to grayscale
retina_gpu = color.rgb2gray(retina_gpu)
# apply four different filters that can enhance vessels
filter_kwargs = dict(sigmas=[2], mode="reflect", black_ridges=True)
filtered_meijering = filters.meijering(retina_gpu, **filter_kwargs)
filtered_sato = filters.sato(retina_gpu, **filter_kwargs)
filtered_frangi = filters.frangi(retina_gpu, **filter_kwargs)
filtered_hessian = filters.hessian(retina_gpu, **filter_kwargs)
The filtered images produced appear as follows:
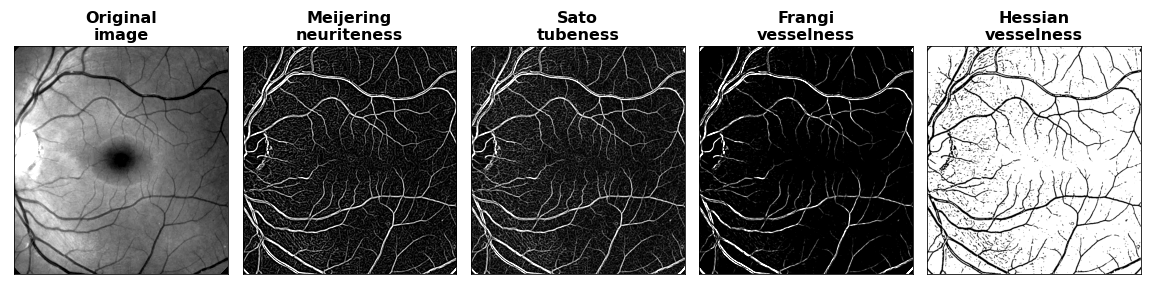
Even for this relatively small-scale image of shape 1011×1011, filtering operations are faster on the GPU than for the corresponding CPU code in scikit-image. In the figure below, acceleration factors relative to scikit-image are shown where the rightmost bar in each group is the acceleration factor observed when round trip host -> device -> host data transfer overhead is included.
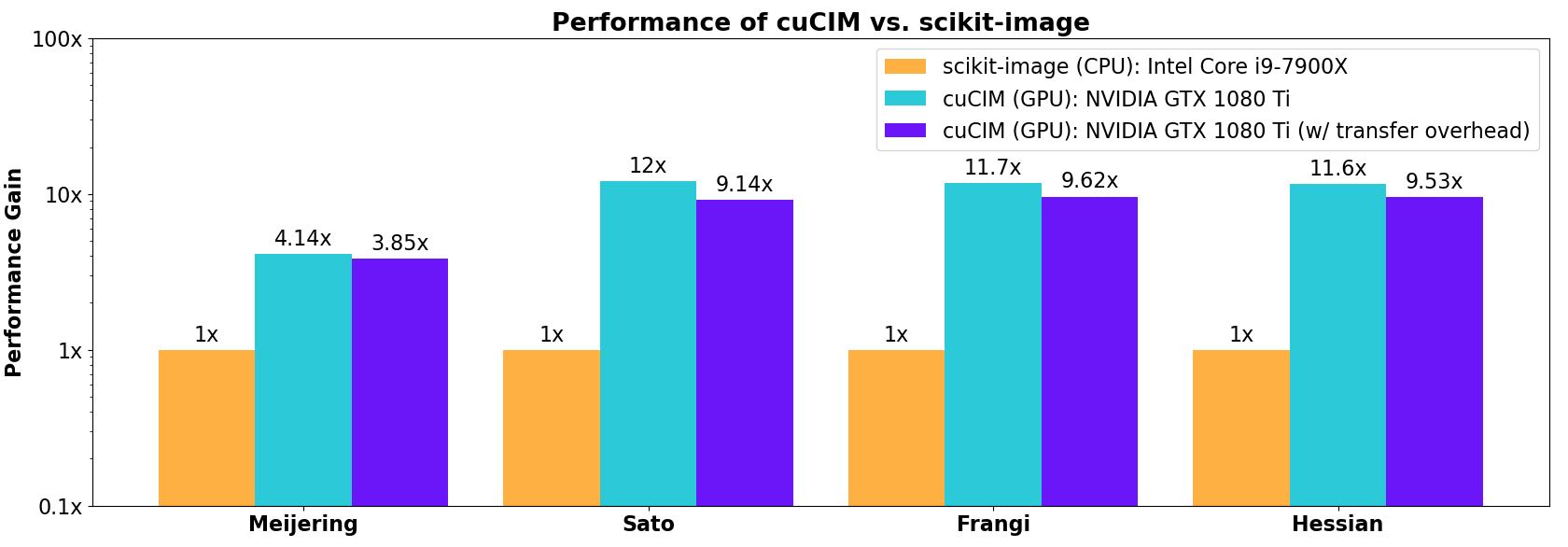
Specific benchmark results across a wider range of functions are highlighted in the companion NVIDIA blog post.
A primary objective of cuCIM is to provide open-source implementations of a wide range of CUDA-accelerated n-dimensional image processing operations that closely mirror the scikit-image API. Volumetric or general n-dimensional data is encountered in scientific fields such as bioimaging (microscopy), medical imaging (CT/PET/MRI), materials science, and remote sensing. This familiar, Pythonic API provides an accessible interface, allowing researchers and data scientists to rapidly port existing CPU-based codes to the GPU.
Although a number of proprietary and open-source libraries provide GPU-accelerated 2D image processing operations (e.g., OpenCV, CUVI, VPI, NPP, DALI), these either lack 3D support or focus on a narrower range of operations than cuCIM. Popular n-dimensional image processing tools like scikit-image, SciPy’s ndimage module, and the Image Processing Toolkit (ITK and SimpleITK) have either no or minimal GPU support. The CLIJ library is an OpenCL-based 2D and 3D image processing library with some overlap in functionality with cuCIM. Although CLIJ is a Java-based project being developed among the ImageJ/Fiji community, an effort is underway to provide interfaces from Python and other languages (clEsperanto).
Aside from providing image processing functions, we also provide image/data readers designed to address the I/O bottlenecks commonly encountered in Deep Learning training scenarios. Both C++ and Python APIs for reading TIFF files with an API matching the OpenSlide library are provided, with support for additional image formats planned for future releases (e.g., DICOM, NIFTI and the Zarr-based Next Generation File Format being developed by the Open Microscopy Environment (OME) project).
cucim.skimage Architecture
The cucim.skimage module is currently implemented using CuPy. CuPy provides a GPU-based implementation of the NumPy API as well as a useful subset of the SciPy API (FFTs, sparse matrices and scipy.ndimage functions). As part of this work we also contributed many image processing functions from the SciPy API back upstream to CuPy itself (see, for example, the image affine transform benchmarks in the complementary NVIDIA post). These capabilities have made it possible to implement a substantial fraction of the scikit-image API in a relatively short time. At this point more than half of the scikit-image API has been implemented and we hope to continue to expand our API coverage over time.
Given that the cucim.skimage module replicates the scikit-image API, it is possible to run many existing scikit-image examples with minimal modification. To demonstrate this, we provide example IPython notebooks that have been adapted from gallery examples. A concrete example involves Gabor filtering on both the CPU and GPU. In this example we have intentionally rerun the same example on both the CPU and GPU, with the only difference between the two cases being the following segment of code:
if use_gpu:
from cupyx.scipy import ndimage as ndi
from cucim.skimage.util import img_as_float32
from cucim.skimage.filters import gabor_kernel
xp = cp
asnumpy = cp.asnumpy
device_name = "gpu"
else:
from scipy import ndimage as ndi
from skimage.util import img_as_float32
from skimage.filters import gabor_kernel
xp = np
asnumpy = np.asarray
device_name = "cpu"
Here it can be seen that the primary difference is in the import statements and the use of a variable xp to represent either the CuPy or NumPy array module. The asnumpy function is defined to transfer data back from the GPU for visualization with Matplotlib. Users inspecting the output on the CPU and GPU will see that it is equivalent and the ratio of the time spent on the CPU vs. GPU will be displayed.
Please note that because CuPy does just-in-time compilation of CUDA kernels the first time they are called, it is expected that the very first time a given example is run, it will have compilation overhead. On repeated executions, previously compiled kernels will be fetched either from an in-memory cache (or from CuPy’s on disk kernel cache in the case of a new Python session).
The necessary changes typically involve:
cupy.asarray to transfer array inputs to cuCIM functions from the host to the GPU.| CPU Module | GPU Module |
|---|---|
numpy | cupy |
scipy | cupyx.scipy |
skimage | cucim.skimage |
sklearn | cuml |
networkx | cugraph |
cupy.asnumpy to transfer results back to the host for plotting with Matplotlib or other visualization libraries or to save results to disk.As is also the case for CuPy itself, cuCIM functions assume that the inputs are already GPU arrays. The user must manage transfer of data to/from the GPU using cupy.asarray or cupy.asnumpy, as appropriate. In general, for best performance, one should try to minimize the number of data transfers between the host and GPU by performing multiple operations on the GPU in sequence. For additional examples see the C++ and Python examples in our repository as well as our GTC conference presentation.
As is also the case for CuPy itself, cuCIM functions assume that the inputs are already GPU arrays. The user must manage transfer of data to/from the GPU using cupy.asarray or cupy.asnumpy, as appropriate. In general, for best performance, one should try to minimize the number of data transfers between the host and GPU by performing multiple operations on the GPU in sequence. For additional examples see the C++ and Python examples in our repository as well as our GTC conference presentation.
cuCIM is an open-source project that welcomes community contributions. We welcome help in reporting bugs, making feature requests or opening pull requests at our GitHub repository. For more details see our Contributor Guidelines. We outline below some areas on our roadmap where we would welcome contributions but are also interested in hearing from the community regarding other ideas. Please reach out and let us know how you are using cuCIM and what areas could use improvement for your application.
Another area of interest is in providing implementations of image processing functions within the C++ API as well. This would not only be of interest to C++ library authors but may also result in implementations that provide improved performance from the Python API.
Another concern is how to keep the existing implementations in sync with upstream scikit-image. Work is starting under a Chan Zuckerberg Institute (CZI) grant with upstream scikit-image to explore ways of dispatching to different computational backends such as cuCIM and dask-image. We aim to avoid unnecessary code duplication where possible and hope to eventually be able to test against different backends to help resolve any discrepancies in behavior among implementations.
We hope to also gradually expand our documentation and tutorials. We would particularly like to highlight instances of the interoperation of cuCIM with other GPU-accelerated packages in the scientific Python ecosystem.
The current 0.19 release of cuCIM uses a newer version of CuPy than other RAPIDS libraries, so installing it alongside these is currently non-trivial. We expect to be fully in sync with other RAPIDS libraries for the upcoming 0.20 release, making it easy to install cuCIM along with other RAPIDS libraries via conda.